Data from: Modeling short visual events through the BOLD Moments video fMRI...
- openneuro.org
Updated Jul 21, 2024 Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied CiteBenjamin Lahner; Kshitij Dwivedi; Polina Iamshchinina; Monika Graumann; Alex Lascelles; Gemma Roig; Alessandro Thomas Gifford; Bowen Pan; SouYoung Jin; N.Apurva Ratan Murty; Kendrick Kay; Radoslaw Cichy*; Aude Oliva* (2024). Modeling short visual events through the BOLD Moments video fMRI dataset and metadata. [Dataset]. http://doi.org/10.18112/openneuro.ds005165.v1.0.4Unique identifierhttps://doi.org/10.18112/openneuro.ds005165.v1.0.4Dataset updatedJul 21, 2024AuthorsBenjamin Lahner; Kshitij Dwivedi; Polina Iamshchinina; Monika Graumann; Alex Lascelles; Gemma Roig; Alessandro Thomas Gifford; Bowen Pan; SouYoung Jin; N.Apurva Ratan Murty; Kendrick Kay; Radoslaw Cichy*; Aude Oliva*License
CiteBenjamin Lahner; Kshitij Dwivedi; Polina Iamshchinina; Monika Graumann; Alex Lascelles; Gemma Roig; Alessandro Thomas Gifford; Bowen Pan; SouYoung Jin; N.Apurva Ratan Murty; Kendrick Kay; Radoslaw Cichy*; Aude Oliva* (2024). Modeling short visual events through the BOLD Moments video fMRI dataset and metadata. [Dataset]. http://doi.org/10.18112/openneuro.ds005165.v1.0.4Unique identifierhttps://doi.org/10.18112/openneuro.ds005165.v1.0.4Dataset updatedJul 21, 2024AuthorsBenjamin Lahner; Kshitij Dwivedi; Polina Iamshchinina; Monika Graumann; Alex Lascelles; Gemma Roig; Alessandro Thomas Gifford; Bowen Pan; SouYoung Jin; N.Apurva Ratan Murty; Kendrick Kay; Radoslaw Cichy*; Aude Oliva*LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
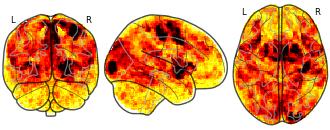
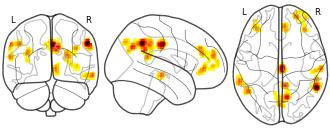
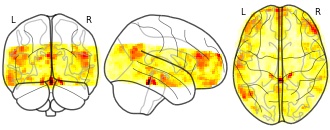
License information was derived automaticallyDescriptionThis is the data repository for the BOLD Moments Dataset. This dataset contains brain responses to 1,102 3-second videos across 10 subjects. Each subject saw the 1,000 video training set 3 times and the 102 video testing set 10 times. Each video is additionally human-annotated with 15 object labels, 5 scene labels, 5 action labels, 5 sentence text descriptions, 1 spoken transcription, 1 memorability score, and 1 memorability decay rate.
Overview of contents:
The home folder (everything except the derivatives/ folder) contains the raw data in BIDS format before any preprocessing. Download this folder if you want to run your own preprocessing pipeline (e.g., fMRIPrep, HCP pipeline).
To comply with licensing requirements, the stimulus set is not available here on OpenNeuro (hence the invalid BIDS validation). See the GitHub repository (https://github.com/blahner/BOLDMomentsDataset) to download the stimulus set and stimulus set derivatives (like frames). To make this dataset perfectly BIDS compliant for use with other BIDS-apps, you may need to copy the 'stimuli' folder from the downloaded stimulus set into the parent directory.
The derivatives folder contains all data derivatives, including the stimulus annotations (./derivatives/stimuli_metadata/annotations.json), model weight checkpoints for a TSM ResNet50 model trained on a subset of Multi-Moments in Time, and prepared beta estimates from two different fMRIPrep preprocessing pipelines (./derivatives/versionA and ./derivatives/versionB).
VersionA was used in the main manuscript, and versionB is detailed in the manuscript's supplementary. If you are starting a new project, we highly recommend you use the prepared data in ./derivatives/versionB/ because of its better registration, use of GLMsingle, and availability in more standard/non-standard output spaces. Code used in the manuscript is located at the derivatives version level. For example, the code used in the main manuscript is located under ./derivatives/versionA/scripts. Note that versionA prepared data is very large due to beta estimates for 9 TRs per video. See this GitHub repo for starter code demonstrating basic usage and dataset download scripts: https://github.com/blahner/BOLDMomentsDataset. See this GitHub repo for the TSM ResNet50 model training and inference code: https://github.com/pbw-Berwin/M4-pretrained
Data collection notes: All data collection notes explained below are detailed here for the purpose of full transparency and should be of no concern to researchers using the data i.e. these inconsistencies have been attended to and integrated into the BIDS format as if these exceptions did not occur. The correct pairings between field maps and functional runs are detailed in the .json sidecars accompanying each field map scan.
Subject 2: Session 1: Subject repositioned head for comfort after the third resting state scan, approximately 1 hour into the session. New scout and field map scans were taken. In the case of applying a susceptibility distortion correction analysis, session 1 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Session 4: Completed over two separate days due to subject feeling sleepy. All 3 testing runs and 6/10 training runs were completed on the first day, and the last 4 training runs were completed on the second day. Each of the two days for session 4 had its own field map. This did not interfere with session 5. All scans across both days belonging to session 4 were analyzed as if they were collected on the same day. In the case of applying a susceptibility distortion correction analysis, session 4 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Subject 4: Sessions 1 and 2: The fifth (out of 5) localizer run from session 1 was completed at the end of session 2 due to a technical error. This localizer run therefore used the field map from session 2. In the case of applying a susceptibility distortion correction analysis, session 1 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Subject 10: Session 5: Subject moved a lot to readjust earplug after the third functional run (1 test and 2 training runs completed). New field map scans were collected. In the case of applying a susceptibility distortion correction analysis, session 5 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
- f
Explaining transient times with moments and tail weight numbers.
- plos.figshare.com
xlsUpdated May 31, 2023 Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied CiteRamakrishnan Iyer; Vilas Menon; Michael Buice; Christof Koch; Stefan Mihalas (2023). Explaining transient times with moments and tail weight numbers. [Dataset]. http://doi.org/10.1371/journal.pcbi.1003248.t002xlsAvailable download formatsUnique identifierhttps://doi.org/10.1371/journal.pcbi.1003248.t002Dataset updatedMay 31, 2023Dataset provided byPLOS Computational BiologyAuthorsRamakrishnan Iyer; Vilas Menon; Michael Buice; Christof Koch; Stefan MihalasLicense
CiteRamakrishnan Iyer; Vilas Menon; Michael Buice; Christof Koch; Stefan Mihalas (2023). Explaining transient times with moments and tail weight numbers. [Dataset]. http://doi.org/10.1371/journal.pcbi.1003248.t002xlsAvailable download formatsUnique identifierhttps://doi.org/10.1371/journal.pcbi.1003248.t002Dataset updatedMay 31, 2023Dataset provided byPLOS Computational BiologyAuthorsRamakrishnan Iyer; Vilas Menon; Michael Buice; Christof Koch; Stefan MihalasLicenseAttribution 4.0 (CC BY 4.0)https://creativecommons.org/licenses/by/4.0/
License information was derived automaticallyDescriptionTable shows the sum of squared residuals for best fit exponentials to (the times taken to reach of the equilibrium firing rate), for the first few moments of 1222 randomly generated synaptic weight distributions between 0 and . For both moments and tail weight numbers, the entries in bold in each column correspond to the lowest value of . Tail weight numbers provide a better fit to transient times than moments.
- N
A shared threat-anticipation circuit is dynamically engaged at different...
- neurovault.org
niftiUpdated Sep 7, 2023+ more versions Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied Cite(2023). A shared threat-anticipation circuit is dynamically engaged at different moments by temporally uncertain and certain threat: CT2mCT3 spmT 0002 [Dataset]. http://identifiers.org/neurovault.image:803231niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:803231Dataset updatedSep 7, 2023License
Cite(2023). A shared threat-anticipation circuit is dynamically engaged at different moments by temporally uncertain and certain threat: CT2mCT3 spmT 0002 [Dataset]. http://identifiers.org/neurovault.image:803231niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:803231Dataset updatedSep 7, 2023LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automaticallyDescriptionCertain-Threat Convolved Block 2 minus Certain-Threat Convolved Block 3 (unthresholded)
Collection description
The Maryland Threat Countdown (MTC) paradigm is a well-established, fMRI-optimized adaptation of temporally uncertain-threat assays that have been behaviorally, pharmacologically, and psychophysiologically validated in rodents and humans. It takes the form of a 2 (Valence: Threat/Safety) × 2 (Temporal Certainty: Certain/Uncertain) repeated-measures, randomized event-related design. We capitalized on the temporally-extended threat-anticipation periods (8.75-30 s, mean = 18.75 s) of the MTC paradigm by modeling temporal dynamics in a more granular manner than before. For continuity with previous research, we performed an initial analysis using a single, variable-length rectangular input function (boxcar) for each anticipation epoch. In our theory-inspired model, we applied three input functions to each anticipation epoch: a brief, transient input function time-locked to anticipation epoch onset, an overlapping variable-length rectangular input function to model sustained activity spanning the entire anticipation epoch and a short, overlapping rectangular input function (6.15 s duration) time-locked to the terminal end of anticipation epochs to capture phasic surges during the ‘circa-strike’ phase. Each of the three input functions was convolved with a canonical HRF. For a second modeling approach, we took a piecewise approach to estimating BOLD magnitude in the temporal domain. For each anticipation period, 2-5 contiguous mini-blocks (6.25 s duration ‘boxcars’) were convolved with a canonical HRF. Findings point to a core neural ‘threat’ system that shows sustained activity across temporally uncertain threat and rapidly assembles in the face of certain-and-imminent threat.
Subject species
homo sapiens
Modality
fMRI-BOLD
Analysis level
group
Cognitive paradigm (task)
rest eyes open
Map type
T
- N
Decoding brain basis of laughter and crying in natural scenes: Second-level...
- neurovault.org
niftiUpdated Jan 30, 2023+ more versions Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied Cite(2023). Decoding brain basis of laughter and crying in natural scenes: Second-level analysis T-map for healthy subjects observing crying in dynamic movie [Dataset]. http://identifiers.org/neurovault.image:790869niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:790869Dataset updatedJan 30, 2023License
Cite(2023). Decoding brain basis of laughter and crying in natural scenes: Second-level analysis T-map for healthy subjects observing crying in dynamic movie [Dataset]. http://identifiers.org/neurovault.image:790869niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:790869Dataset updatedJan 30, 2023LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automaticallyDescriptionCollection description
Subject species
homo sapiens
Modality
fMRI-BOLD
Analysis level
group
Cognitive paradigm (task)
emotional localizer fMRI task paradigm
Map type
T
- N
Motivational signals disrupt metacognitive signals in the human ventromedial...
- neurovault.org
niftiUpdated Feb 1, 2022+ more versions Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied Cite(2022). Motivational signals disrupt metacognitive signals in the human ventromedial prefrontal cortex: GLM2b choice moment pmod early certainty [Dataset]. http://identifiers.org/neurovault.image:776245niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:776245Dataset updatedFeb 1, 2022License
Cite(2022). Motivational signals disrupt metacognitive signals in the human ventromedial prefrontal cortex: GLM2b choice moment pmod early certainty [Dataset]. http://identifiers.org/neurovault.image:776245niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:776245Dataset updatedFeb 1, 2022LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automaticallyDescriptionCollection description
Subject species
homo sapiens
Modality
fMRI-BOLD
Analysis level
group
Cognitive paradigm (task)
self monitoring task
Map type
T
- N
Task-invariant networks interfere with and task-specific networks support...
- neurovault.org
niftiUpdated May 21, 2025+ more versions Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied Cite(2025). Task-invariant networks interfere with and task-specific networks support memory formation: An fMRI meta-analysis: Low-quality > High-quality encoding, pictorial tasks [Dataset]. http://identifiers.org/neurovault.image:900169niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:900169Dataset updatedMay 21, 2025License
Cite(2025). Task-invariant networks interfere with and task-specific networks support memory formation: An fMRI meta-analysis: Low-quality > High-quality encoding, pictorial tasks [Dataset]. http://identifiers.org/neurovault.image:900169niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:900169Dataset updatedMay 21, 2025LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automaticallyDescriptionCollection description
Why do some moments imprint themselves in memory while others vanish without a trace? This meta-analysis uncovers a marked dissociation in the brain’s large-scale networks during memory encoding: networks that impede encoding are largely task-invariant, whereas those that support it are finely tuned to the task at hand. Drawing on fMRI studies using the subsequent memory paradigm, the analysis contrasts neural activity during the encoding of later-remembered versus later-forgotten trials across verbal and pictorial tasks. Leveraging Yeo et al.’s 17-network parcellation and a novel network association metric, the results show that memory-impeding effects consistently recruit the same set of subsystems within the default mode, frontoparietal, and ventral attention networks across different tasks, indicating a shared neural signature of distraction or mind-wandering. In contrast, memory-supporting effects exhibit clear task-dependent divergence: verbal encoding engages language-related networks, while pictorial encoding activates visuo-perceptual systems. This asymmetry suggests that forgetting arises from similar internal mechanisms across contexts, whereas successful encoding depends on precise, context-sensitive neural engagement. These findings highlight the dynamic interplay between memory and attention and provide a network-level perspective on how the brain toggles between remembering and forgetting.
Subject species
homo sapiens
Modality
fMRI-BOLD
Analysis level
meta-analysis
Cognitive paradigm (task)
encoding task
Map type
R
- N
Processing of visual and non-visual naturalistic spatial information in the...
- neurovault.org
niftiUpdated Mar 1, 2021+ more versions Share
Share Facebook
Facebook Twitter
Twitter EmailClick to copy linkLink copied
EmailClick to copy linkLink copied Cite(2021). Processing of visual and non-visual naturalistic spatial information in the "parahippocampal place area": AO contrast 10 (vse_new, vpe_new > vse_old, vpe_old) [Dataset]. http://identifiers.org/neurovault.image:407565niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:407565Dataset updatedMar 1, 2021License
Cite(2021). Processing of visual and non-visual naturalistic spatial information in the "parahippocampal place area": AO contrast 10 (vse_new, vpe_new > vse_old, vpe_old) [Dataset]. http://identifiers.org/neurovault.image:407565niftiAvailable download formatsUnique identifierhttps://identifiers.org/neurovault.image:407565Dataset updatedMar 1, 2021LicenseCC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automaticallyDescriptioncontrol at moments of cuts
Collection description
Subject species
homo sapiens
Modality
fMRI-BOLD
Analysis level
group
Cognitive paradigm (task)
audio narrative
Map type
Z
Not seeing a result you expected?
Learn how you can add new datasets to our index.
 Facebook
Facebook Twitter
TwitterData from: Modeling short visual events through the BOLD Moments video fMRI dataset and metadata.
CC0 1.0 Universal Public Domain Dedicationhttps://creativecommons.org/publicdomain/zero/1.0/
License information was derived automatically
This is the data repository for the BOLD Moments Dataset. This dataset contains brain responses to 1,102 3-second videos across 10 subjects. Each subject saw the 1,000 video training set 3 times and the 102 video testing set 10 times. Each video is additionally human-annotated with 15 object labels, 5 scene labels, 5 action labels, 5 sentence text descriptions, 1 spoken transcription, 1 memorability score, and 1 memorability decay rate.
Overview of contents:
The home folder (everything except the derivatives/ folder) contains the raw data in BIDS format before any preprocessing. Download this folder if you want to run your own preprocessing pipeline (e.g., fMRIPrep, HCP pipeline).
To comply with licensing requirements, the stimulus set is not available here on OpenNeuro (hence the invalid BIDS validation). See the GitHub repository (https://github.com/blahner/BOLDMomentsDataset) to download the stimulus set and stimulus set derivatives (like frames). To make this dataset perfectly BIDS compliant for use with other BIDS-apps, you may need to copy the 'stimuli' folder from the downloaded stimulus set into the parent directory.
The derivatives folder contains all data derivatives, including the stimulus annotations (./derivatives/stimuli_metadata/annotations.json), model weight checkpoints for a TSM ResNet50 model trained on a subset of Multi-Moments in Time, and prepared beta estimates from two different fMRIPrep preprocessing pipelines (./derivatives/versionA and ./derivatives/versionB).
VersionA was used in the main manuscript, and versionB is detailed in the manuscript's supplementary. If you are starting a new project, we highly recommend you use the prepared data in ./derivatives/versionB/ because of its better registration, use of GLMsingle, and availability in more standard/non-standard output spaces. Code used in the manuscript is located at the derivatives version level. For example, the code used in the main manuscript is located under ./derivatives/versionA/scripts. Note that versionA prepared data is very large due to beta estimates for 9 TRs per video. See this GitHub repo for starter code demonstrating basic usage and dataset download scripts: https://github.com/blahner/BOLDMomentsDataset. See this GitHub repo for the TSM ResNet50 model training and inference code: https://github.com/pbw-Berwin/M4-pretrained
Data collection notes: All data collection notes explained below are detailed here for the purpose of full transparency and should be of no concern to researchers using the data i.e. these inconsistencies have been attended to and integrated into the BIDS format as if these exceptions did not occur. The correct pairings between field maps and functional runs are detailed in the .json sidecars accompanying each field map scan.
Subject 2: Session 1: Subject repositioned head for comfort after the third resting state scan, approximately 1 hour into the session. New scout and field map scans were taken. In the case of applying a susceptibility distortion correction analysis, session 1 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Session 4: Completed over two separate days due to subject feeling sleepy. All 3 testing runs and 6/10 training runs were completed on the first day, and the last 4 training runs were completed on the second day. Each of the two days for session 4 had its own field map. This did not interfere with session 5. All scans across both days belonging to session 4 were analyzed as if they were collected on the same day. In the case of applying a susceptibility distortion correction analysis, session 4 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Subject 4: Sessions 1 and 2: The fifth (out of 5) localizer run from session 1 was completed at the end of session 2 due to a technical error. This localizer run therefore used the field map from session 2. In the case of applying a susceptibility distortion correction analysis, session 1 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.
Subject 10: Session 5: Subject moved a lot to readjust earplug after the third functional run (1 test and 2 training runs completed). New field map scans were collected. In the case of applying a susceptibility distortion correction analysis, session 5 therefore has two sets of field maps, denoted by “run-1” and “run-2” in the filename. The “IntendedFor” field in the field map’s identically named .json sidecar file specifies which functional scans correspond to which field map.